The goal of this blogpost is to present a concise implementation of the Gaussian Mixture Model (GMM) using einsum notation. Along the way, I will also describe the expectation-maximization (EM) algorithm.
Einsum
Einstein notation is a particular convention of writing tensor operations that is implemented in NumPy, PyTorch and other deep learning frameworks. Its essence lies in indicating summations implicitly: the summation is over indices that are skipped or occur repeatedly. For instance, a batched bilinear transformation \(d = aBc\), for \(a \in \mathbb{R}^{B \times I}, B \in \mathbb{R}^{B \times I \times J}, c \in \mathbb{E}^{B \times J}\), can be wrriten as \(d_b = a_i B_{ij} c_j\) or np.einsum('bi,bij,bj->b', a, B, c). As I will show below, it is particularly useful when dealing with mixtures or multivariate Gaussians, which are effectively 3-dimensional tensors.
Tim Rocktäschel has a nice introductory post on einsum in the context of deep learning.
Gaussian mixtures
Let’s assume we have a dataset \(X\) of \(N\) data points in an \(D\)-dimensional space. Here \(D=2\) and \(N=100\).
def create_dataset():
x1 = np.random.normal(size=(100, 2)) + np.array([-1, -1])
x2 = np.random.normal(size=(100, 2)) + np.array([1, -1])
x3 = np.random.normal(size=(100, 2)) + np.array([0, 1])
return np.vstack((x1, x2, x3))
X = create_dataset()
plt.scatter(X[:, 0], X[:, 1], cmap='gist_rainbow', s=50)
sns.despine()
 |
|---|
| The dataset \(X \in \mathbb{R}^D\) for our motivating example |
We would like to discover clusters in the data and assign each point \(x_n \in X\) with a categorical probability distribution over \(K\) clusters. To that end, we will try to model the dataset as a mixture of \(K\) Gaussians.
Gaussian mixtures can be posed as latent variable models of the following form:
\[p_\theta(X,Z) = \prod_{n=1}^N \prod_{k=1}^K \Big[\pi_k \mathcal{N}(x_n|\mu_k, \Sigma_k) \Big]^{z_{nk}},\]where the first product is over data points and the second over mixture components. \(z_{nk}\) is a latent variable determining whether \(n\)-th data point belongs to \(k\)-th mixture components and the parameters are \(\theta = \{\pi_k, \mu_k, \Sigma_k\}_k\).
Will will implement our GMM as a class GaussianMixture with a scikit-learn-like API:
class GaussianMixture:
def __init__(self, num_components, input_size, max_iter=10):
self.num_components = num_components
self.input_size = input_size
self.max_iter = max_iter
# Parameters
self.pi = np.ones((self.num_components))/self.num_components
self.mu = np.random.normal(size=(self.num_components, self.input_size))
self.sigma = np.tile(np.eye(self.input_size), (self.num_components, 1, 1))
When fitting the model, we are interested in maximising the logarithm of \(p_\theta(X,Z)\):
\[\log p(X,Z) = \sum_{n=1}^N \sum_{k=1}^K z_{nk} \Big[\log \pi_k + \log \mathcal{N}(x_n|\mu_k, \Sigma_k) \Big].\]To make this problem, we instead maximise the following lower bound
\[\mathcal{L}(\theta, q) = \sum_{n=1}^N \sum_{k=1}^K q(z|x_n) \Big[\log \pi_k + \log \mathcal{N}(x_n|\mu_k, \Sigma_k) - \log q(z|x)\Big]\]where \(q(z \vert x)\) is a variational distribution introduced for convenience: a categorical over \(K\) components. \(\mathcal{L}\) is is known as the ELBo or evidence lower bound as it is always the case that \(\mathcal{L} \leq p_\theta(X)\).
The EM algorithm
EM is an algorithm for maximum likelihood (or maximum a posteriori) estimation of parameters for latent variable models. The algorithm is iterative: we alternate between minimising \(\mathcal{L}(\theta, q)\) with respect to \(\theta\) and updating \(q\) to make the bound tighter, each time fixing one and updating the other. We to that until \(\mathcal{L}\) stops increasing.
def fit(self, X):
prev_elbo = -np.inf
for i in range(self.max_iter):
q = self._expect(X)
self.pi, self.mu, self.cov = self._maximize(X, q)
elbo = self.compute_elbo(X, q)
if np.allclose(elbo, prev_elbo):
break
prev_elbo = elbo
E-step
In the E-step, we infer the \(q(z \vert x)\) that inverts \(p_\theta(x \vert z)\) for current \(\theta\) as determined by Bayes’ theorem. For a particular data point \(n\) and a component \(k\), we have:
\[q(z_k|x_n) = \frac{p(z_k)p(x_n|z_k)}{p(x_n)} = \frac{\pi_k \mathcal{N}(x_n|\mu_k, \Sigma_k)}{\sum_{k'=1}^K \pi_{k'} \mathcal{N}(x_n|\mu_{k'}, \Sigma_{k'})}\]def _expect(self, X):
n = X.shape[0]
c = self.num_components
d = self.input_size
pi = self.pi.reshape((1, c))
X = X.reshape((n, 1, d))
mu = self.mu.reshape((1, c, d))
inv_sigma = np.linalg.inv(self.sigma)
det_sigma = np.linalg.det(self.sigma)
distance = np.einsum('ncd,cde,nce->nc', X - mu, inv_sigma, X - mu)
Z = np.sqrt(det_sigma) * (2 * np.pi) ** (d/2)
p_x_given_t = np.einsum('nc,c->nc', np.exp(-distance/2), 1/Z)
q_unnormalized = p_x_given_t * pi
return q_unnormalized/q_unnormalized.sum(axis=1, keepdims=True)
M-step
In the M-step, we maximise \(\mathcal{L}\) with respect to \(\theta\) given \(q(z \vert x)\) from the E-step. There are closed-form solutions for each of the parameters:
\[\pi_k = \frac{1}{N}\sum_{n=1}^N z_{nk},\] \[\mu_k = \frac{1}{N_k} \sum_{n=1}^N q(z_k|x_n)x_n,\] \[\Sigma_k = \frac{1}{N_k} \sum_{n=1}^N q(z_k|x_n) (x_n-\mu_k) (x_n-\mu_k)^{\text T},\]where \(N_k = \sum_{n=1}^N q(z_k \vert x_n)\) can be interpreted as a pseudo-count of data points assigned to component \(k\).
def _maximize(self, X, q):
n = X.shape[0]
c = self.num_components
d = self.input_size
nk = q.sum(axis=0).reshape((c, 1))
mu = np.einsum('nd,nc->cd', X, q)/nk
error = X.reshape((n, 1, d)) - mu.reshape((1, c, d))
sigma = np.einsum('ncd,nce,nc->cde', error, error, q)/nk.reshape(c, 1, 1)
pi = (nk/n).reshape((c,))
return pi, mu, sigma
Computing the ELBo
Finally, a we also need to compute the ELBo which we use as a stopping criterion (although in principle it could also be used as a validation metric or a debugging tool). Recall that the ELBo is just
\[\mathcal{L}(\theta, q) = \sum_{n=1}^N \sum_{k=1}^K q(z|x_n) \Big[\log \pi_k + \log \mathcal{N}(x_n|\mu_k, \Sigma_k) - \log q(z|x)\Big].\]def compute_elbo(self, X, q):
n = X.shape[0]
c = self.num_components
d = self.input_size
q += 10e-20
pi = self.pi.reshape((1, c))
X = X.reshape((n, 1, d))
mu = self.mu.reshape((1, c, d))
inv_sigma = np.linalg.inv(self.sigma)
logdet_sigma = np.linalg.slogdet(self.sigma)[1].reshape(1, c)
distance = np.einsum('ncd,cde,nce->nc', X - mu, inv_sigma, X - mu)
constants = -d/2*np.log(2*np.pi)
log_p_x_given_t = constants - logdet_sigma/2 - distance.reshape((n, c))/2
log_pi = np.log(pi).reshape((1, c))
log_q = np.log(q)
return ((log_pi + log_p_x_given_t - log_q) * q).sum()
Using the model
Since our latent \(q(z_k \vert z_n)\) is interpretable as cluster assignments, clustering the data boils down to running the E-step with a trained model:
def predict(self, X):
return self._expect(X)
Let us illustrate the obtained components by colour-coding soft cluster assignments:
gmm = GaussianMixture(num_components=3, input_size=2)
gmm.fit(X)
y = gmm.predict(X)
plt.scatter(X[:, 0], X[:, 1], c=y, cmap='gist_rainbow', s=50)
sns.despine()
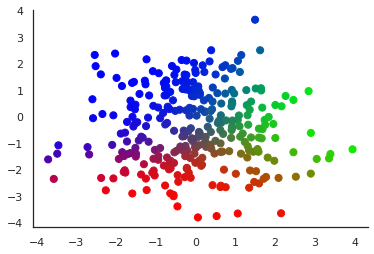 |
|---|
| The dataset \(X\) with each \(x_n\) displayed in a colour obtained from interpreting the vector \(q(z \vert x_n)\) as an RGB triplet: \(z_{n1}\) codes for the value of the red component, \(z_{n2}\) for green and \(z_{n3}\) for blue |
EM and the wake-sleep algorithm
There is a non-trivial connection between the EM algorithm and the wake-sleep algorithm I described earlier. The wake step corresponds to the M-step: minimizing the ELBo w.r.t. \(p_\theta\). The sleep step corresponds to the E-step with an important quirk: the input is not an \(x\) sampled from the dataset \(X\), but a synthetic example sampled from \(p(x \vert z)\), where the \(z\) is in turn sampled from a prior \(p(z)\). Moreover, the \(q(z \vert x)\) is an amortised model: it is parametrised by some weights that are should minimise the ELBo across data points.
A notebook accompanying this blog post is available on GitHub. It also contains some tests of my einsum implementation against a simpler one with an explicit for loop over \(K\) components and calls to scipy.stats.multivariate_normal.
